Biological interpretation
To illustrate the performance of TTSMI database, we employ TTSMI database to the analysis of an application of triplex technology such as modulation of gene expression of TNF-α (Paquet et al. 2011) and IGF1 (Trojan et al. 2010 and 2012) .
Examples
TNF-α
Here, we provide an example of gene regulation on TNF-α via antigene technology using TFO recognition of the TTS located within the promoter region of the gene. Gene regulation via TFO recognition has been largely exploited in order to modulate gene expression. In studies of in vitro and in vivo systems, TFOs have been shown to modulate gene expression during the transcription process, by interfering with the binding of either the transcription factors or the transcription initiation complex formation to the target DNA (Duca et al. 2008 and Jain et al. 2010). One of the studies using this approach was examined for tumor necrosis factor-α (TNF-α) gene (Paquet et al. 2011). An anti-TNF-α triplex-forming oligonucleotide (TFO) could target the TTS located within the promoter region of TNF-α gene (rn4, chr20:3660925-3660945, -54 bases from TSS, 5'-TCGAAAAGGGGTGGGAGAAGG-3') and modulate its gene expression, resulting in the reduction of disease development in both acute and chronic rat arthritis models.
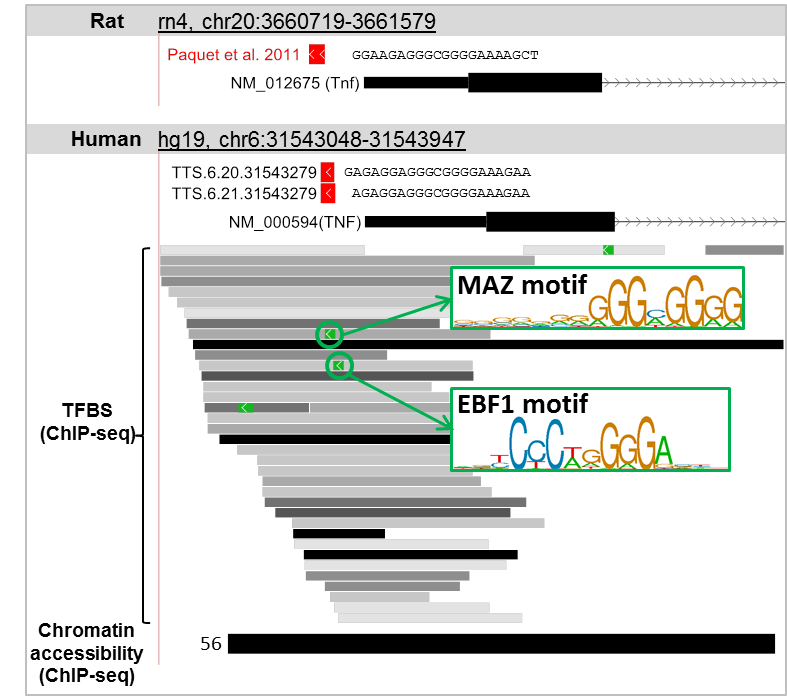
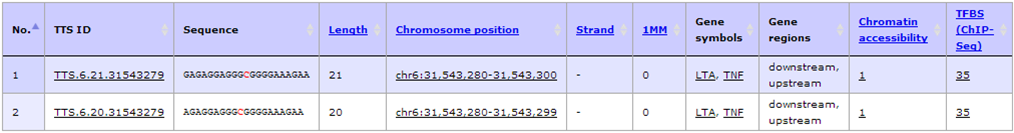
Using TTSMI database to search for TTSs in TNF gene promoter with defined parameter settings (more than 40% G content, 1 pyrimidine interruption, number of GpGpG less than 4, TTS off-target site of 1 mismatch equal to zero, and the TTS overlapping with "Putative promoter", "Chromatin accessibility (ChIP-seq)", and "TFBS (ChIP-seq)"), we identified two TTSs that passed the filtering criteria including TTS.6.21.31543279 (hg19, chr6:31543280-31543300, -43 bases from TSS, 5'-GAGAGGAGGGCGGGGAAAGAA-3') and TTS.6.20.31543279 (hg19, chr6:31543280-31543299, -44 bases from TSS, 5'- AGAGGAGGGCGGGGAAAGAA-3').
Interestingly, these TTSs were found to be conserved between the human and rat genomes (as shown in figure below). In addition, these TTSs co-localize with chromatin accessibility regions defined by ChIP-seq in 56 cell-types, 35 TFBS defined by ChIP-seq, and 2 TFBS motifs in MAZ and EBF1 binding regions. This evidence suggests that both TTSs (TTS.6.21.31543279 and TTS.6.20.31543279) could be good candidates for further investigation as part of a therapeutic tool for TNF-a-dependent inflammatory disorders in human.
Below is a step-by-step instruction on how to find the TTS described above.
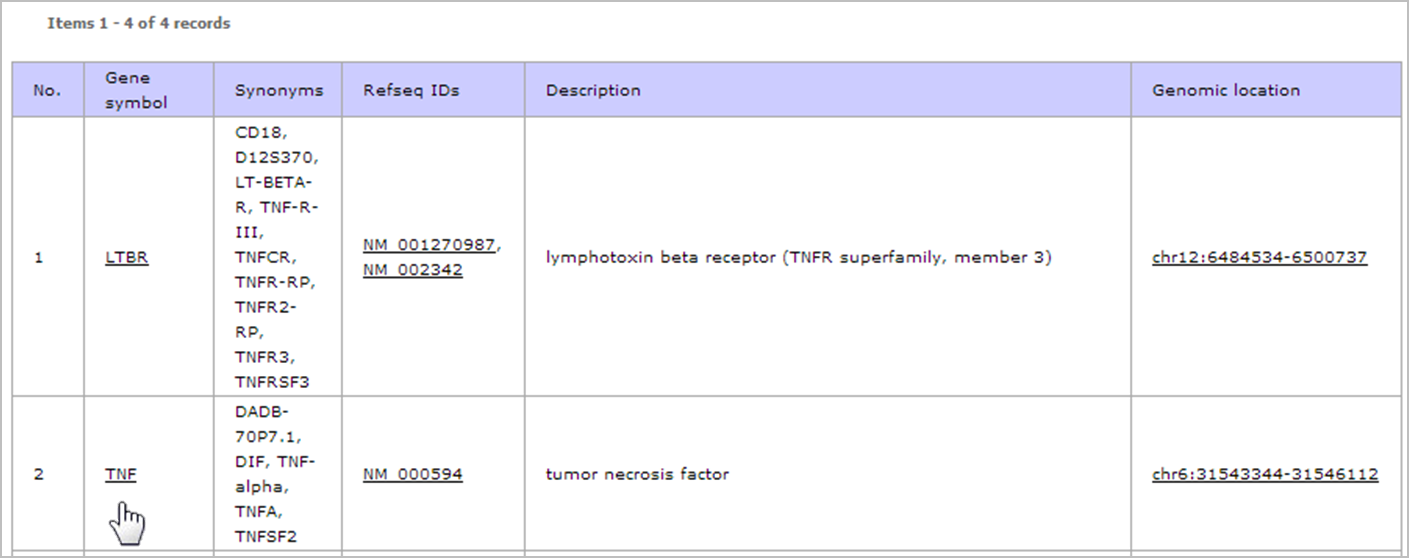
- Type gene name or gene description keywords of interest in the search term box. In this example, we are looking for TTS of TNF gene. So we type "TNF" in the search term box and press enter.
- The result shows list of genes that match to "TNF". Then click on the gene name (TNF) at "Gene symbol" column to view the TTS that are located on the gene.
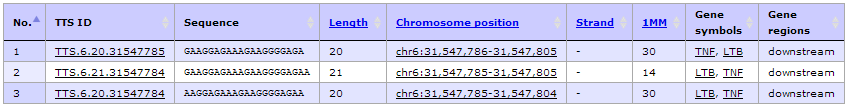
- Unfortunately, the default setting of TTS search criteria shows a list of 3 records of TTSs that are located on downstream regions of TNF gene. Indeed, we would like to find TTSs that could be used as antigene technology as well as it is able to regulate TNF gene expression by interfering the binding of transcription factor binding sites.
-
Next we then adjust the searching criteria and narrow down the list of TTSs with the highest potential for gene regulation by filtering with several criteria.
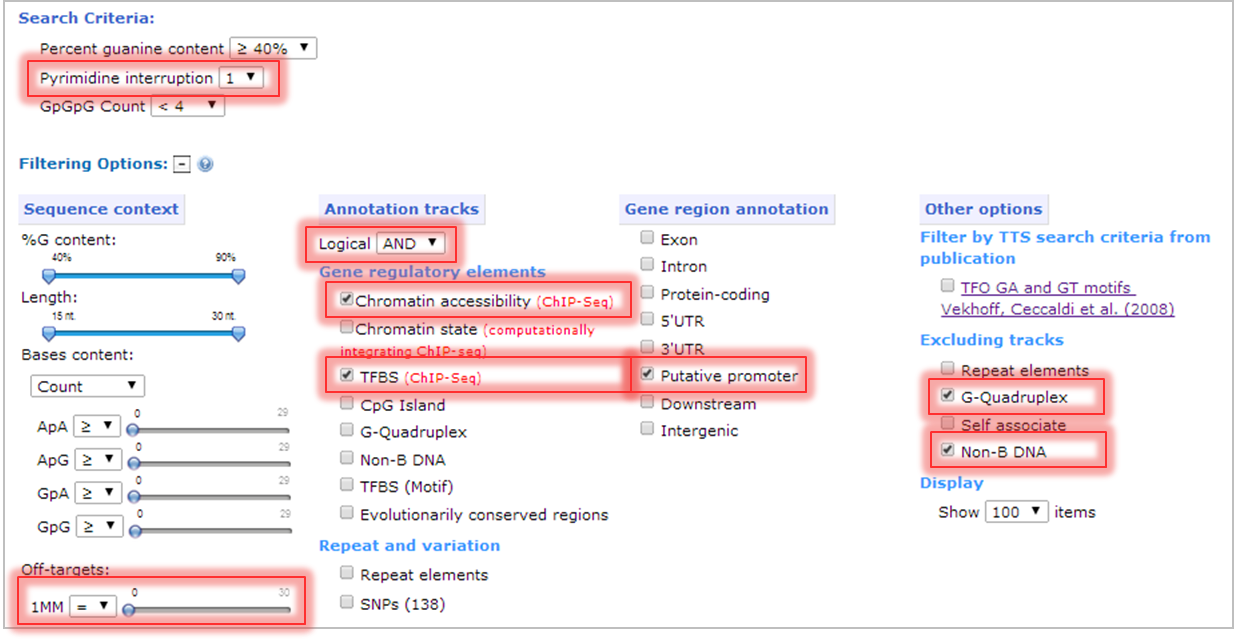
- TTS search criteria: allow 1 pyrimidine interruption.
- Limit number of potential TTS off-target sites: number of potential TTS off-target sites with 1 mismatch equal to zero.
- Annotation tracks: select chromatin accessibility (ChIP-seq) and TFBS (ChIP-Seq) to filter TTS that belong to open chromatin structure and transcription factor binding sites. The logical operator is set as "AND" to select the TTS that co-localize with both annotation tracks.
- Gene region annotation: select Putative promoter to filter TTS that belong to promoter region.
- Excluding alternative DNA structure: select G-quadruplex and Non-B DNA sequence that might able to interfere the formation of triplex structure.
- Click submit!
- The result shows only two TTS left including TTS.6.21.31543279 (hg19, chr6:31543280-31543300, -43 bases from TSS, 5'-GAGAGGAGGGCGGGGAAAGAA-3') and TTS.6.20.31543279 (hg19, chr6:31543280-31543299, -44 bases from TSS, 5'- AGAGGAGGGCGGGGAAAGAA-3'). Then click at "TTS ID" column to view more complete annotation of a given TTS.
IGF1
Here, we provide an example of an application of antigene technology in clinical trials. It has been shown that TFO (e.g. anti-IGF-I) can effectively stop the development of animal tumours and human gliomas and malignant tumours (Trojan et al. 2010 and 2012). Moreover, it has been reported that antisense anti-IGF-I has been used in Phase I and II of clinical trials to patients with glioblastoma and other types of cancers (Trojan et al. 2012). These successful cases of using antigene technology should be considered as promising models for specific antigene therapy of cancer and other medical conditions, and other applications.
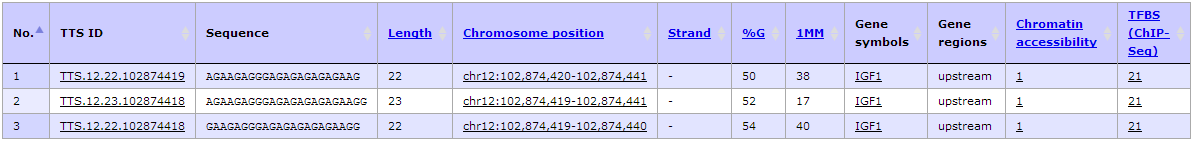
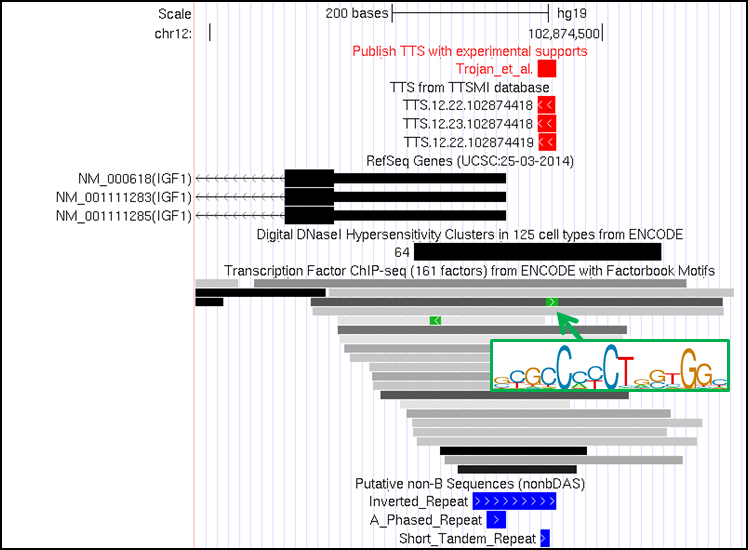
IGF-I (type I insulin-like growth factor) is one of the most important growth factors related to cell and tissue differentiation and proliferation. The inhibition of IGF1 expression leads to reduce tumorigenicity of cells in mouse and human. It has been reported that anti-IGF-I approach can be used for inhibiting transcription of igf1 gene via TFO-based technology in both mouse and human tumours (Trojan et al. 2010 and 2012). An anti-IGF-I TFO could target the TTS located within the promoter region of IGF1 gene (hg19, chr12:102874419-102874441, -40 bases from TSS, 5'-AGAAGAGGGAGAGAGAGAGAAGG-3') and modulate its gene expression, resulting in the reduction of tumours development.
Using TTSMI database to search for TTSs in IGF1 gene promoter with defined parameter settings (more than 40% G content, no pyrimidine interruption, number of GpGpG less than 4, filtering criteria by Vekhoff et al. 2008 and the TTS overlapping with "Putative promoter", "Chromatin accessibility (ChIP-seq)", and "TFBS (ChIP-seq)"), we identified three TTSs that passed the filtering criteria including TTS.12.22.102874419 (hg19, chr12:102874420-102874441, -41 bases from TSS, 5'-AGAAGAGGGAGAGAGAGAGAAG-3'), TTS.12.23.102874418 (hg19, chr12:102874419-102874441, -40 bases from TSS, 5'-AGAAGAGGGAGAGAGAGAGAAGG-3'), and TTS.12.22.102874418 (hg19, chr12:102874419-102874440, -40 bases from TSS, 5'-GAAGAGGGAGAGAGAGAGAAGG-3').
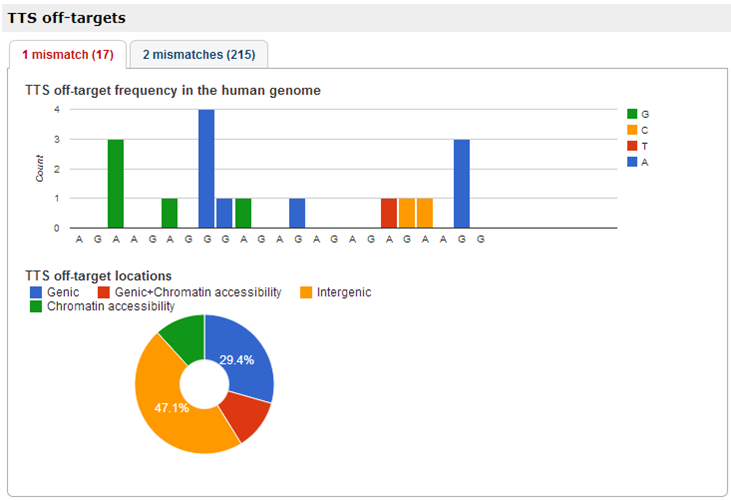
We find the previously used target site (Trojan et al. 2010 and 2012 i.e. TTS.12.23.102874418) among three TTSs (as shown in figure below). In addition, these TTSs co-localize with chromatin accessibility regions defined by ChIP-seq in 64 cell-types, 21 TFBS defined by ChIP-seq, and 1 TFBS motifs in CTCF binding region. Although this evidence suggests that these TTSs might be good candidates for further investigation as part of a therapeutic tool, the potential TTS off-target sites of the anti-IGF-I TFO should carefully address. Since we found that, with 1 mismatch, anti-IGF-I TFO could potential forming triplex in 17 locations of human genome. More than half (9 out of 17 sites) of the potential off-targets might form triplex in genic region and/or chromatin accessibility regions.
Below is a step-by-step instruction on how to find the TTS described above.
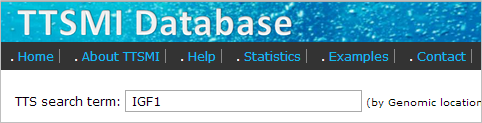
- Type gene name or gene description keywords of interest in the search term box. In this example, we are looking for TTS of IGF1 gene. So we type "IGF1" in the search term box and press enter.
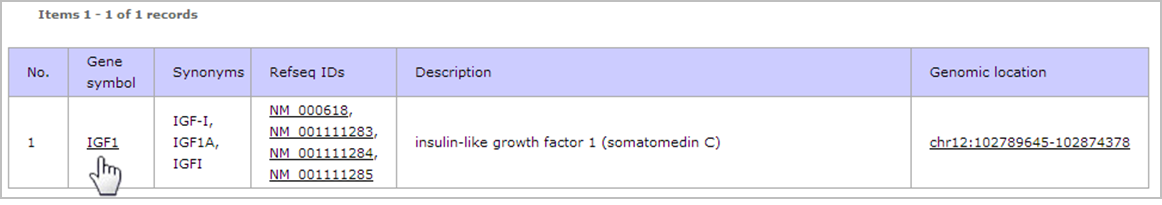
- The result shows list of genes that match to "IGF1". Then click on the gene name (IGF1) at "Gene symbol" column to view the TTS that are located on the gene.
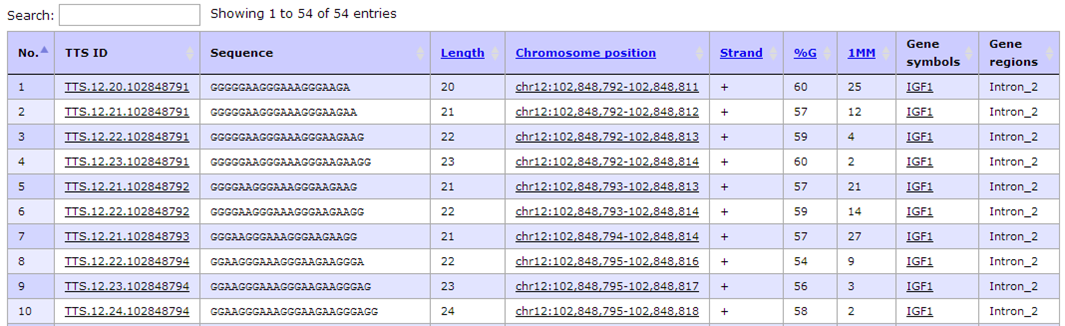
- Unfortunately, the default setting of TTS search criteria shows a list of 54 records of TTSs that are located on intronic and upstream regions of IGF1 gene. Indeed, we would like to find TTSs that could be used as antigene technology as well as it is able to regulate IGF1 gene expression by interfering the binding of transcription factor binding sites.
-
Next we then adjust the searching criteria and narrow down the list of TTSs with the highest potential for gene regulation by filtering with several criteria.
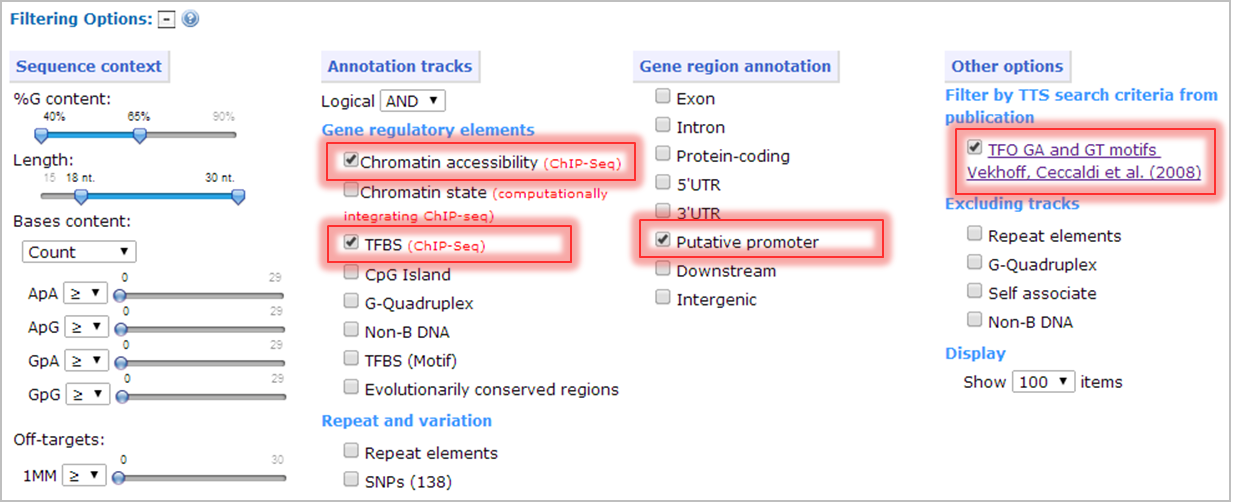
- Annotation tracks: select chromatin accessibility (ChIP-seq) and TFBS (ChIP-Seq) to filter TTS that belong to open chromatin structure and transcription factor binding sites. The logical operator is set as "AND" to select the TTS that co-localize with both annotation tracks.
- Gene region annotation: select Putative promoter to filter TTS that belong to promoter region.
- Filtering based on TTS search criteria from publication: select TTS as following criteria: (i) length of the TTS range from 18 to 30 nt, (ii) %G content of the TTS range from 40 to 65%, (iii) the stretches of guanines are not longer than 4 nt, (iv) the poly-adenines are not longer than 7 nt, (v) the number of stretches of three guanines does not exceed 3, and (vi) the TTS is unique in the human genome.
- Click submit!
- The result shows only three TTS left including TTS.12.22.102874419 (hg19, chr12:102874420-102874441, -41 bases from TSS, 5'-AGAAGAGGGAGAGAGAGAGAAG-3'), TTS.12.23.102874418 (hg19, chr12:102874419-102874441, -40 bases from TSS, 5'-AGAAGAGGGAGAGAGAGAGAAGG-3'), and TTS.12.22.102874418 (hg19, chr12:102874419-102874440, -40 bases from TSS, 5'-GAAGAGGGAGAGAGAGAGAAGG-3'). Then click at "TTS ID" column to view more complete annotation of a given TTS.
- The potential TTS off-target sites of the anti-IGF-I TFO could be viewed after step 5. We found that, with 1 mismatch, anti-IGF-I TFO could potentially form triplex structure in 17 locations of human genome. More than half (9 out of 17 sites) of the potential off-targets might form triplex in genic and/or chromatin accessibility regions. Although, TFO could potentially form triplex structure to 17 off-target sites, the stability of the TFO and the off-target sites is much less than the TTS target (Mergny, 1991 and Roberts, 1991). This report could help the researcher to further investigate non-target triplex-forming and could control off-target effects for each TTS.